Deposition
DeePiCt Training and Evaluation Datasets
- Deposition ID:CZCDP-10000
Release Date: 2023-06-01
Last Modified: 2023-06-01
Deposition Overview
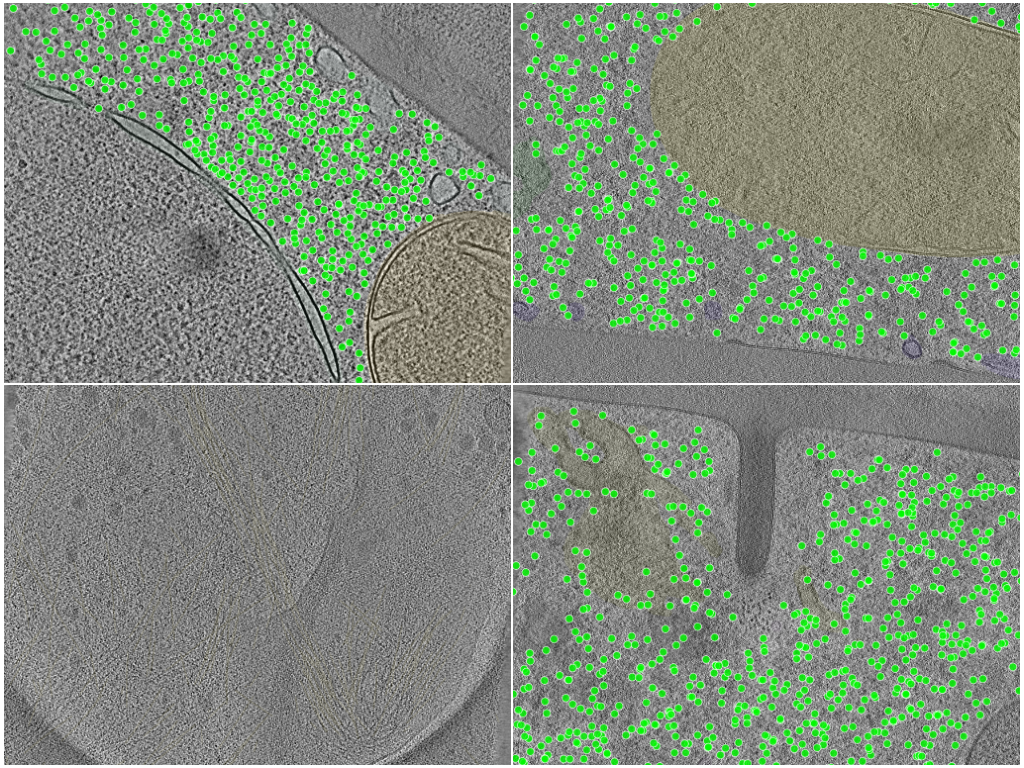
Datasets comprising 20 annotated Schizosaccharomyces pombe cryo-electron tomograms, with 10 tomograms acquired using a Volta phase plate (VPP) for enhanced contrast and 10 using conventional defocus. The dataset includes comprehensive annotations of ribosomes, fatty acid synthases, membranes, organelles, and cytosol. Additionally, 3 tomograms of RPE-1 cells used to train networks for actin segmentation are included. These datasets enabled the development and validation of DeePiCt, a deep learning framework combining 2D and 3D convolutional neural networks for cellular structure segmentation and macromolecular complex localization across different species and acquisition conditions.
Authors
Irene de Teresa Trueba, Sara Goetz, Alexander Mattausch, Frosina Stojanovska, Christian Eugen Zimmerli, Mauricio Toro-Nahuelpan, Dorothy W. C. Cheng, Fergus Tollervey, Constantin Pape, Martin Beck, Alba Diz-Munoz, Anna Kreshuk, Julia Mahamid, Judith B. Zaugg
Annotation Methods Summary
Method Type
Hybrid
Method Links
Not Submitted
Datasets with Deposition Data
3 of 3 Datasets
Dataset Name | Organism | Runs With deposition data | Annotations Deposition only | Annotated Objects Deposition only | |
|---|---|---|---|---|---|
 | S. pombe cells from Volta Phase Plate Dataset ID: DS-10001 Irene de Teresa Trueba, ... , Julia Mahamid, Judith B. Zaugg | Schizosaccharomyces pombe | 10 | 86 |
|
 | RPE1 cytosol with actin stress fiber Dataset ID: DS-10002 Irene de Teresa Trueba, ... , Julia Mahamid, Judith B. Zaugg | Homo sapiens | 3 | 6 |
|
 | Dataset ID: DS-10000 Irene de Teresa Trueba, ... , Julia Mahamid, Judith B. Zaugg | Schizosaccharomyces pombe | 10 | 83 |
|